
This article summarizes major researches that applied graph neural networks (GNN) to analyzing electronic health records (EHR).
GRAM: Graph-based Attention Model
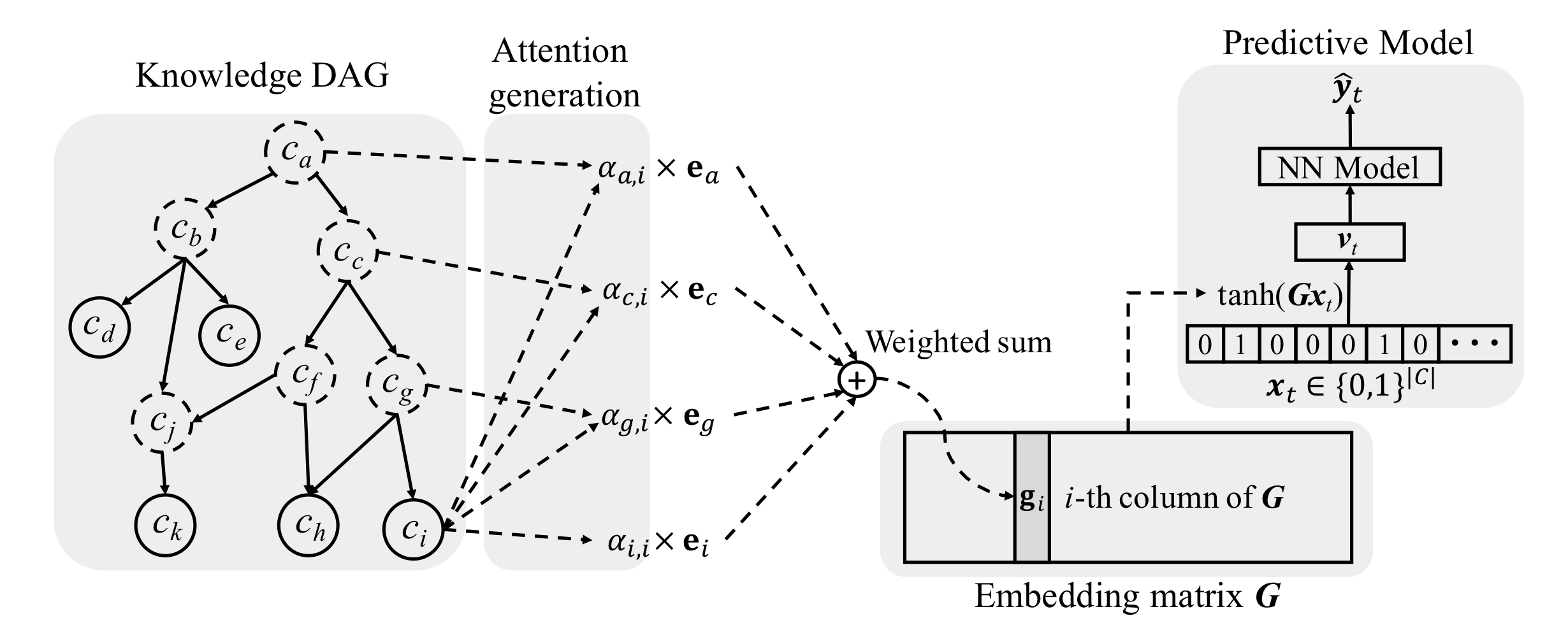
Image source: (Choi, Bahadori, Song, Stewart, & Sun, 2017)
Notations:
(Choi, Bahadori, Song, Stewart, & Sun, 2017) infused medical ontologies to deep learning models through neural attention. In this paper, ontology
Node embeddings (
where
Comments
They exploited the parent-child relationship to update embeddings but completely forfeited hierarchical structure of the graph. The attention calculation adopted did not distinguish parents from different levels, which contained valuable information because for DAG, directed connected nodes are intuitively more similar than nodes
Authors used co-occurrence information to initialize node embeddings. Co-occurrence favors more general concepts, which occurs more frequently across patients and thus, , more general and less discriminative. From this perspective, the representations of general concepts may not be as “reliable” as claimed in the paper. Additionally, information infiltrated from a higher-level concept tends to make the representations of all its leaf nodes uniform, making the leaf node representation indiscriminative. The co-occurrence matrix was sorted by ancestors (see the figure below). It is unclear where to put node b in the matrix because it is both the first ancestor and the third ancestor.
The co-occurrence matrix was sorted by ancestors (see the figure below). It is unclear where to put node
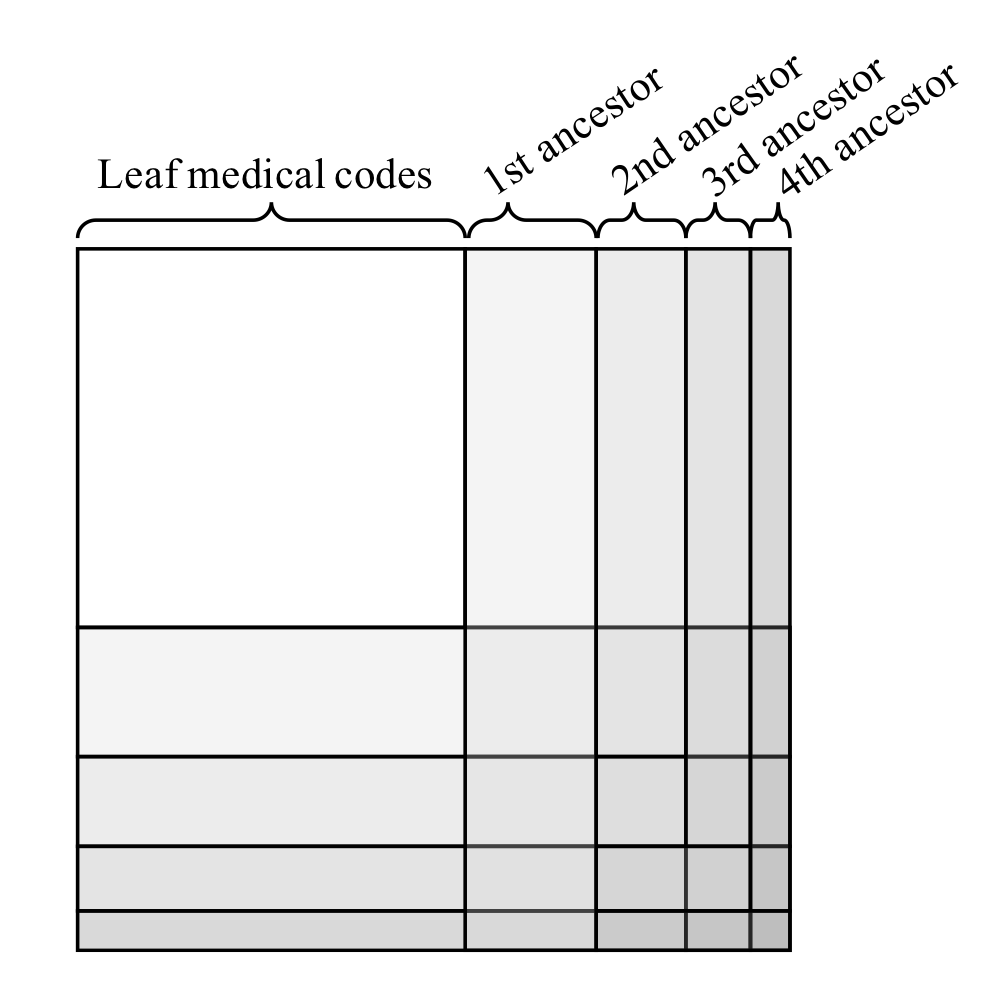
Image source: (Choi, Bahadori, Song, Stewart, & Sun, 2017)
MiME: Multilevel Medical Embedding
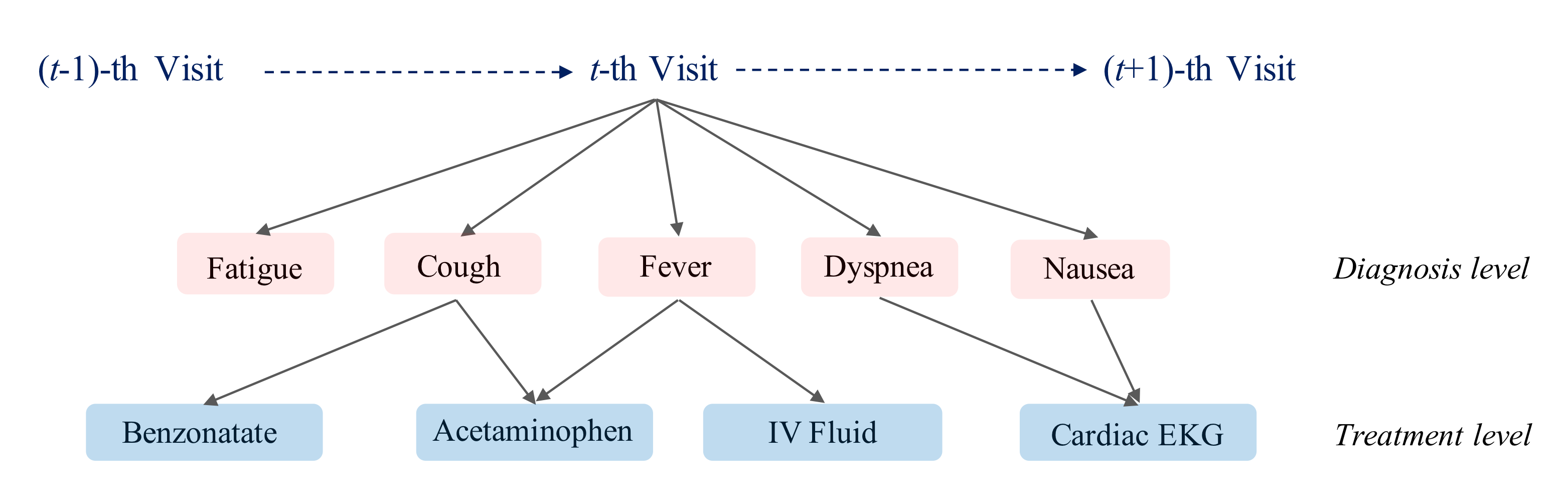
Image source: (Choi, Xiao, Stewart, & Sun, 2018)
In this paper (Choi, Xiao, Stewart, & Sun, 2018), authors exploited the underlying EHR structure rather than standard medical ontologies. The graph was defined by four hierarchical levels: patient, visit, diagnosis and treatment. In MiME, the interaction between diagnosis and treatment was captured by element-wise multiplication.
where
Graph Convolutional Transformer for EHR
(Choi et al., 2020) proposed a way of extracting visit representations with implicit graph structures of medical concepts. (If the graph is explicitly defined, MiME (Choi et al., 2018) can be applied to solve the problem.) Authors assumed that all concepts were fully connected, and attention layers from a trained transformer were able to assign higher weights to meaningful connections. In the figure below, when converged, the model assigned higher weights (thicker arrow) to more meaningful connections.
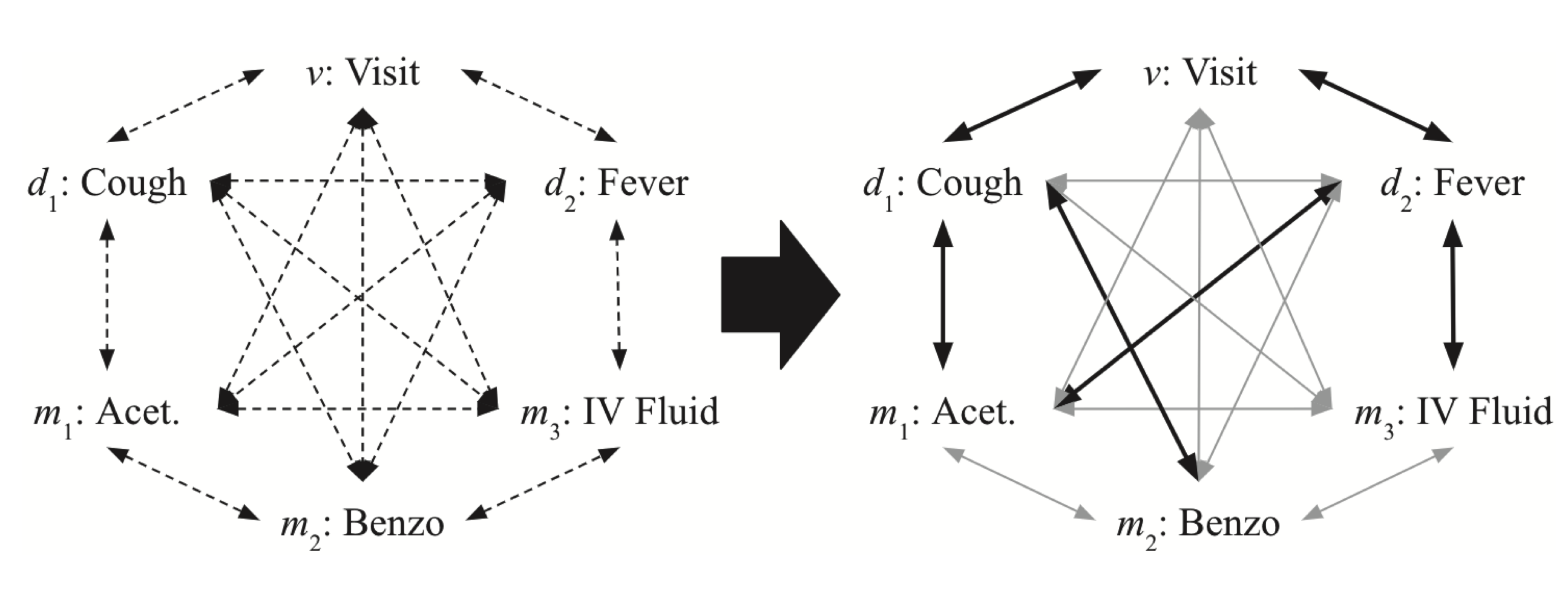
Image source: (Choi et al., 2020)
Without guidance, the model must search the entire attention space. Authors constrained the search space by imposing two rules
- Masking connections that are not allowed by medical facts
- Replace the attention mechanism of the first layer with the conditional distribution among concepts and penalize the divergence of attention weights in higher layers from the conditional distribution.
Comments
Framed as a graph convolutional transformer, the model is not very different from a vanilla transformer encoder. It is essentially a transformer encoder with attention weight distribution guided and special masks applied. In addition, the attention weights are assumed to be able to be directly used as interpretation (i.e., feature similarities/connections).
MedGCN
Authors (Mao, Yao, & Luo, 2019) developed a model that imputed laboratory test results and gave medication recommendations to patients. They considered four types of medical entities: encounters, patients, labs and medications and their relations.
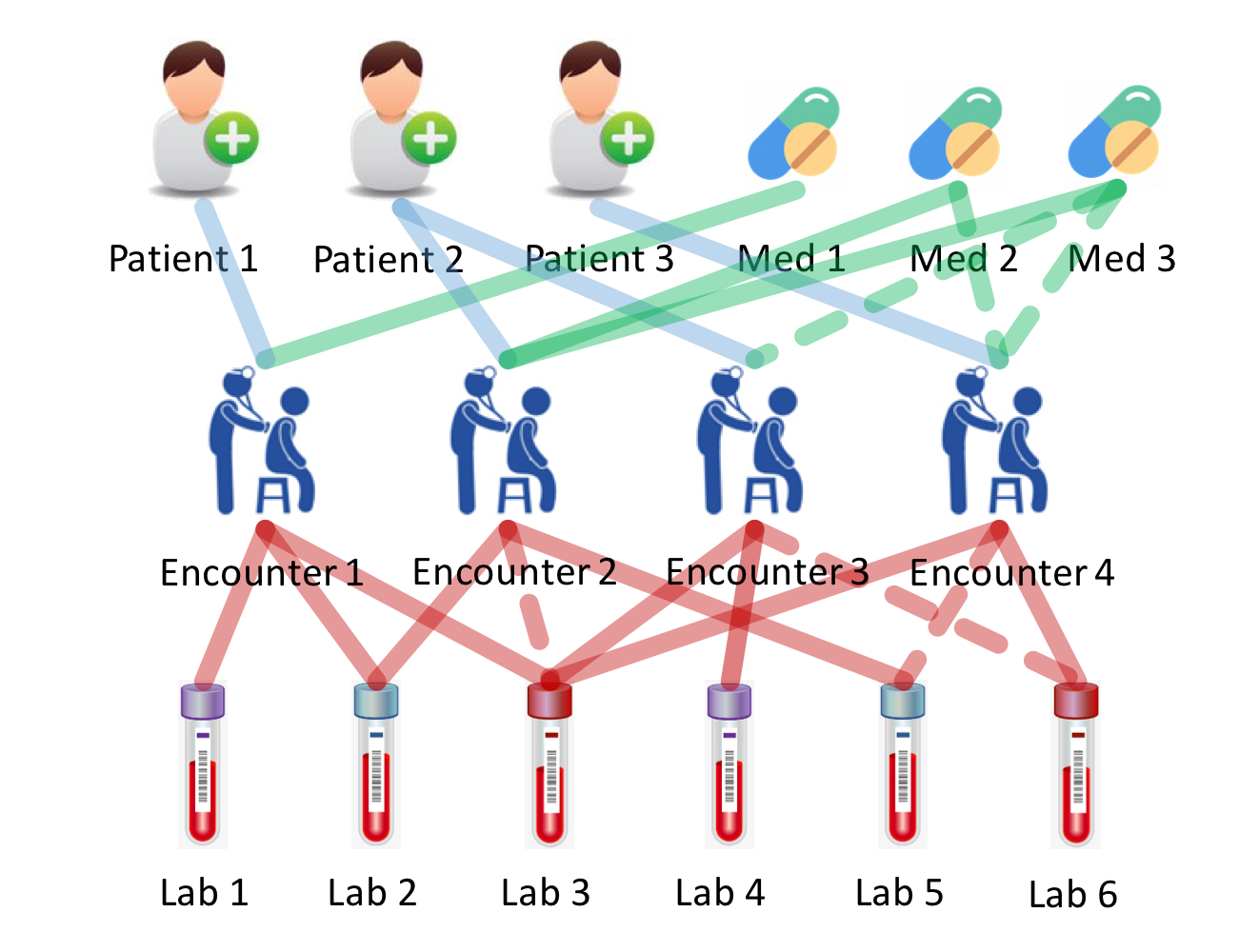
Solid lines represent observed relations and dashed lines indicate the relations are unknown. Image source: (Mao, Yao, & Luo, 2019)
The relations between encounters and patients, as well as encounters and medications were encoded as binary adjacency matrices, while those between lab tests and encounters were encoded in a sparse continuous matrix, with nonzero entries being the normalized lab tests results. They created an additional mask matrix to distinguish “real 0” from “missing 0. Encounters served as the central concepts that connect to all other concepts. With the help of these defined matrices, they updated all node following the standard GCN update rules, except different weights were used for connections between different concepts.
They built two independent deep learning models that mapped the encounter representations to two prediction targets (medication recommendations and lab tests imputations) respectively. Massage passing and two predictive models were trained end-to-end. The final loss function is the weighted sum of losses of two tasks, which they called “cross regularization”. The total loss only regulated the node representation updates but did not regulate the training of two tributary models.
Comments
The research question was ill-posed for two reasons. First, medications information was infiltrated to the encounter representations and the model in turn tried to predict medications from encounter representations. Second, based on the task formulation, model recommended medications only after it had access to all information associated to that encounter, which was equivalent to proposing disease intervention methods when patients were about to be discharged. I suspect the defined graph structure only had very marginal contribution to the model performance, because the connections were purely co-occurrence (the “belongs to” relation). The events co-occurrence is modelled implicitly in all models that do not operate on graphs.
HeteroMed
This (Hosseini, Chen, Wu, Sun, & Sarrafzadeh, 2018) was the first work to use Heterogeneous Information Network (HIN) for modeling clinical data and disease diagnosis. HIN is a graph whose nodes and/or edges are of various types.
A clinical event e is defined as
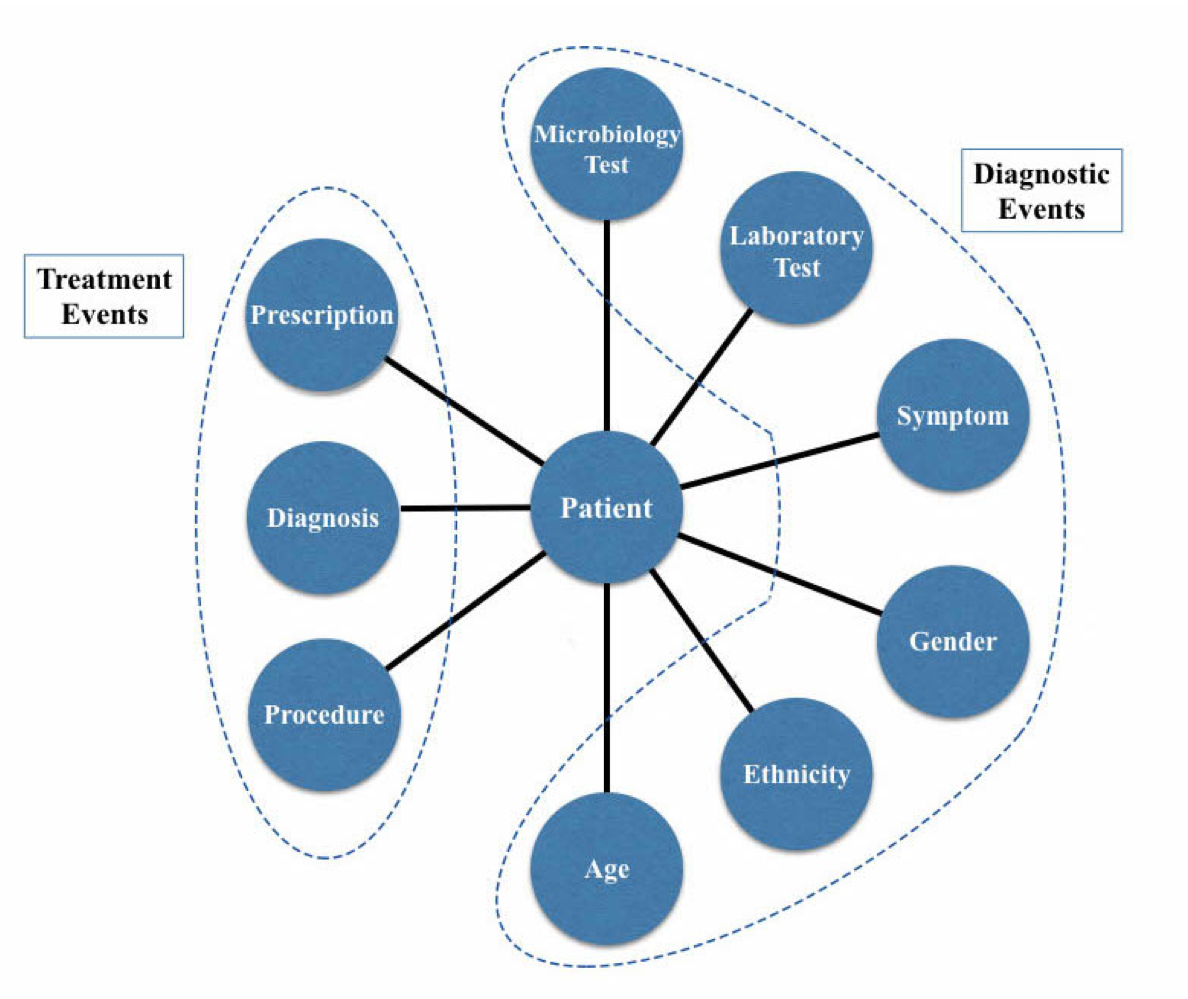
EHR heterogeneous network schema. Image source: (Hosseini, Chen, Wu, Sun, & Sarrafzadeh, 2018)
Links in this work indicated the “belong to” relation. In addition to the “belong to” relation derived from EHR, authors also defined meta paths (Chang et al., 2015) because they were believed to be able to better learn the semantics of similarity among nodes. For instances, patient
To learn representation of nodes (the unsupervised task in this paper), they devised a task of predicting the observed neighborhood of a node
where
The computation of
where
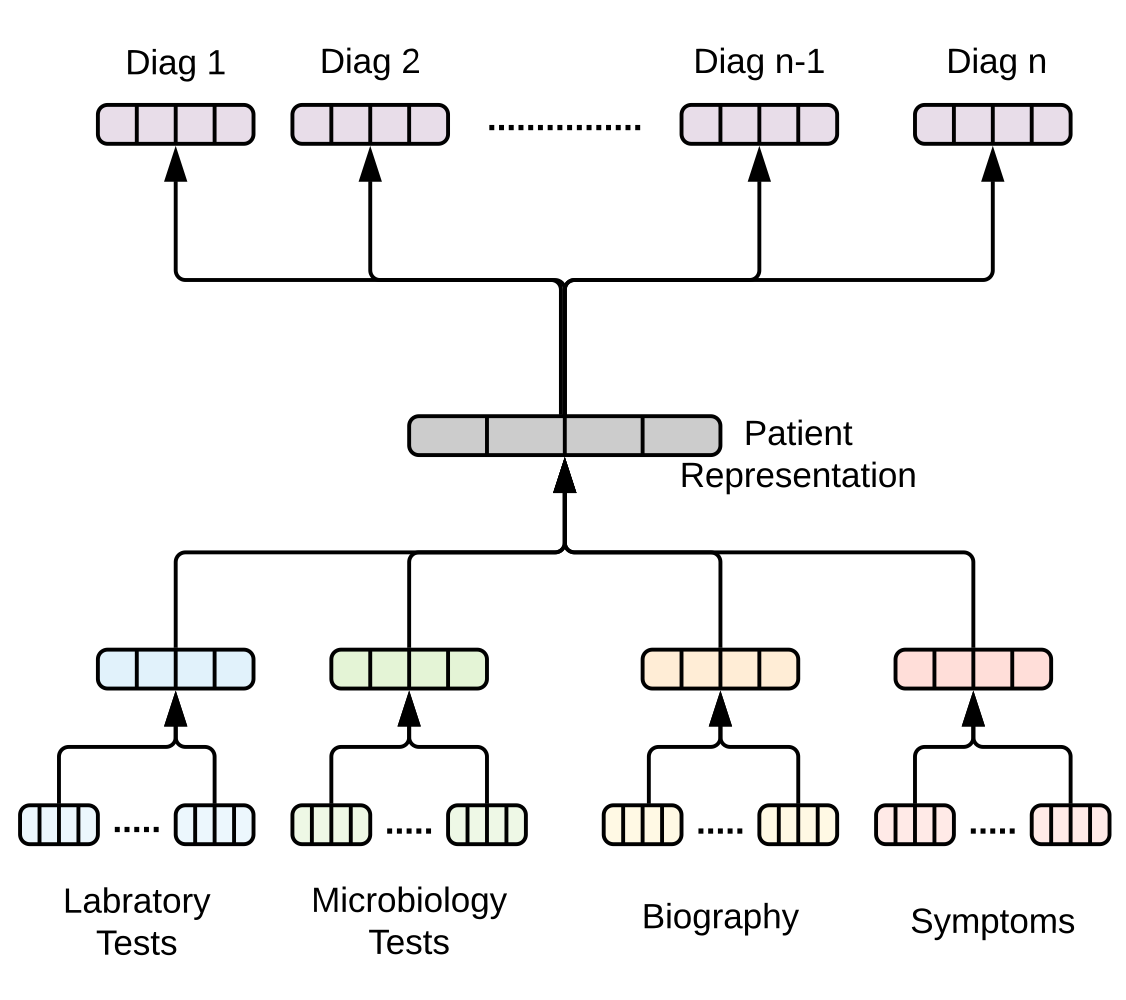
Diagnosis prediction flow. Image source: (Hosseini, Chen, Wu, Sun, & Sarrafzadeh, 2018)
To predict diagnoses of a patient (the supervised task in this paper), they framed the problem as follows. Given a patient
A representation of a patient
where the weights
Finally, a diagnosis
They subsequently employed the hinge loss ranking objective for the triple
To add guidance for learning representations that specific for the diagnosis task, inspired by (Chen & Sun, 2017), they jointly trained the supervised and unsupervised task, i.e., jointly updated embedding parameters and model weights by defining an objective
where
Comments
Authors used almost full spectrum of event types in EHR and disentangled them into diagnosis types and treatment types. The study integrated many advanced techniques into its modeling pipeline, such as Autophrase (Shang et al., 2018) to extract symptom phrases and meta paths (Chang et al., 2015) to impose reasonable bias, and is an excellent and inspiring work.
The study design was rigorous in the sense that the model predicted diagnoses based on the diagnostic events. However, since treatment events were involved in updating the embeddings of diagnostic events (authors declared that prescription was used to update representations of symptom but did not point out if diagnosis was used to update any representations of diagnostic events), due to information infiltration along the graph paths, treatment events (prescription, diagnosis and procedure) were implicitly used to predict diagnoses.
Furthermore, since the model’s predictions were based on complete episodes, the model had no incentive or probably incapable of deriving timely predictions. This further limits its utility. Additionally, due to doctor’s intervention, patients’ conditions might change over time, this could plague the embedding learning and the prediction. One simple solution may be modeling only adverse events rather than all of them.
HealGCN

Overall flowchart of HealGCN. Source: (Wang et al., 2021)
Authors developed a system that allowed users to self-diagnoze their diseases (Wang et al., 2021). The pipeline consisted of three steps: step 1, question and answer; step 2: inference for diagnosis; and step 3: diagnosis results display. Only the second step is relevant to our discussion, therefore, included in this blog.
They constructed a heterogenous graph based on three groups of concepts from EHR: symptom
where
The trainable embedding matrix only contains disease and symptoms embeddings. User embeddings are not included because the number of users was too large, so maintaining a large user embedding matrix was not tractable. Additionally, most users were cold started, the user embedding matrix would not be sufficiently trained to be representative. User representations were generated on the fly based on neighboring symptom nodes.
They also included the concept of meta paths: Disease-Symptom-Disease (DSD) and User-Symptom-User (USU). Given a meta path
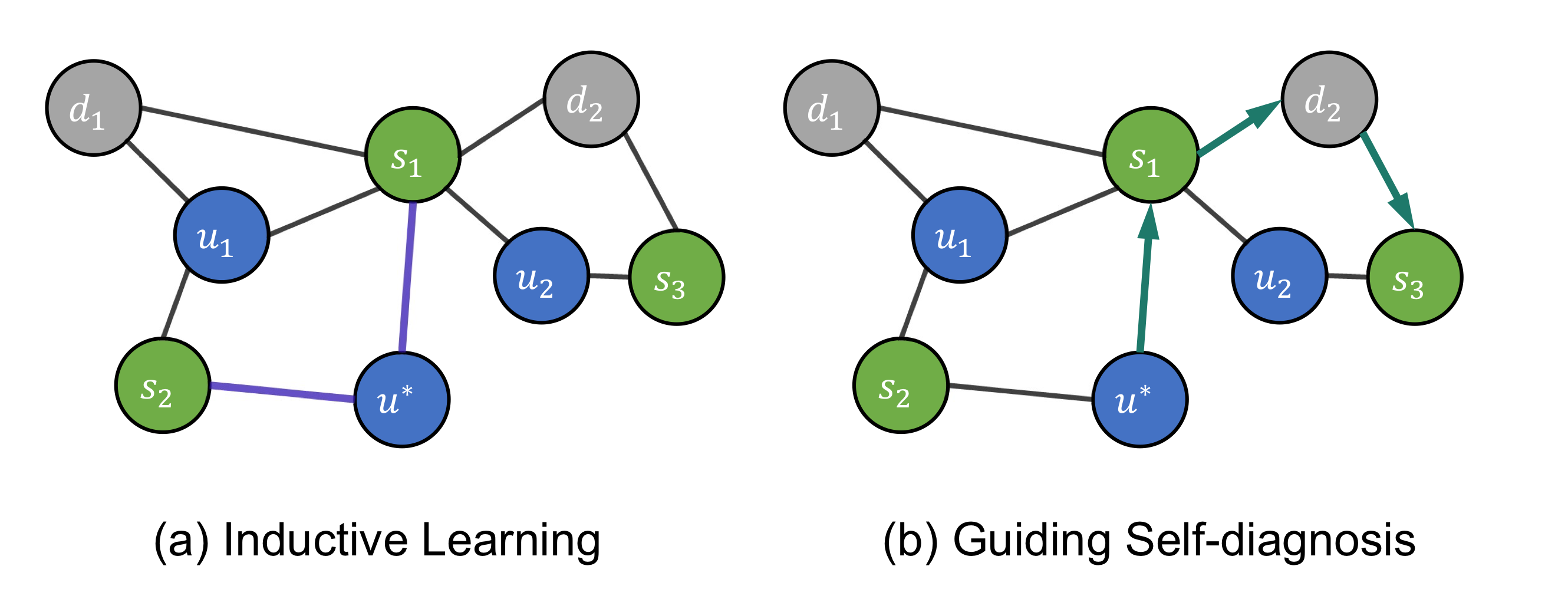
Source: (Wang et al., 2021)
The paper adopted 1-hop message passing and followed the standard GCN message passing rules except when transforming the neighbor representations. Instead of being a simple linear mapping
where
The paper adopted Bayesian Personalized Ranking (BPR) loss (Rendle, Freudenthaler, Gantner, & Schmidt-Thieme, 2012)
where
Comments
Authors made multiple innovative model designs, including mega paths (Chang et al., 2015; Hosseini et al., 2018), message passing formula, and the loss function. The model allowed some node representations to be extracted on the fly, solving the problems of cold-start users, improving scalability inductive power. However, these innovations and their rationale were not discussed in depth. Their true utility calls for further investigation.
References
- Chang, S., Han, W., Tang, J., Qi, G.-J., Aggarwal, C. C., & Huang, T. S. (2015). Heterogeneous network embedding via deep architectures. Paper presented at the Proceedings of the 21th ACM SIGKDD international conference on knowledge discovery and data mining.
- Chen, T., & Sun, Y. (2017). Task-guided and path-augmented heterogeneous network embedding for author identification. Paper presented at the Proceedings of the Tenth ACM International Conference on Web Search and Data Mining.
- Choi, E., Bahadori, M. T., Song, L., Stewart, W. F., & Sun, J. (2017). GRAM: graph-based attention model for healthcare representation learning. Paper presented at the Proceedings of the 23rd ACM SIGKDD international conference on knowledge discovery and data mining.
- Choi, E., Xiao, C., Stewart, W. F., & Sun, J. (2018). Mime: Multilevel medical embedding of electronic health records for predictive healthcare. arXiv preprint arXiv:1810.09593.
- Choi, E., Xu, Z., Li, Y., Dusenberry, M., Flores, G., Xue, E., & Dai, A. (2020). Learning the graphical structure of electronic health records with graph convolutional transformer. Paper presented at the Proceedings of the AAAI Conference on Artificial Intelligence.
- Hosseini, A., Chen, T., Wu, W., Sun, Y., & Sarrafzadeh, M. (2018). Heteromed: Heterogeneous information network for medical diagnosis. Paper presented at the Proceedings of the 27th ACM International Conference on Information and Knowledge Management.
- Mao, C., Yao, L., & Luo, Y. (2019). Medgcn: Graph convolutional networks for multiple medical tasks. arXiv preprint arXiv:1904.00326.
- Rendle, S., Freudenthaler, C., Gantner, Z., & Schmidt-Thieme, L. (2012). BPR: Bayesian personalized ranking from implicit feedback. arXiv preprint arXiv:1205.2618.
- Shang, J., Liu, J., Jiang, M., Ren, X., Voss, C. R., & Han, J. (2018). Automated phrase mining from massive text corpora. IEEE Transactions on Knowledge and Data Engineering, 30(10), 1825-1837.
- Wang, Z., Wen, R., Chen, X., Cao, S., Huang, S.-L., Qian, B., & Zheng, Y. (2021). Online Disease Diagnosis with Inductive Heterogeneous Graph Convolutional Networks. Paper presented at the Proceedings of the Web Conference 2021.
- Post title:Graph Neural Networks and Electronic Health Records
- Post author:Lutao Dai
- Create time:2022-01-15 14:43:00
- Post link:https://lutaodai.github.io/2022-01-15-gnn-ehr/
- Copyright Notice:All articles in this blog are licensed under BY-NC-SA unless stating additionally.